Bacterial Transformation
Escherichia coli are commensal gram negative bacteria found in the guts of humans. They have the capacity to double every twenty minutes and make a favorable carrier of recombinant DNA. Plasmid DNA can be introduced into E. coli easily after making them competent. One method to achieve this is through chemical competence with heat shock. In this process, the bacteria are incubated in CaCl2 solution on ice. The cold serves to slow down molecular motion of the plasma membrane while the Ca2+ ions remove the charge-charge repulsion between the phospholipids and the negatively charged DNA seeking to gain entry into the cell. Cells are placed for a short period of time at 42°C to induce heat shock. This heat shock results in the cell taking up the DNA. This method is very low efficiency so many bacteria do not take in any DNA. Cells are allowed to recover from heat shock at 37°C in rich nutrient broth to allow for the production of the antibiotic resistance proteins encoded on the vector as a selection marker. Transformed cells are then spread across an agar plate containing the antibiotic which will then kill all non-transformed cells. Only the bacteria containing the vector with the antibiotic resistance gene will survive and replicate to form small colonies on the surface of the agar.
- http://www.dnalc.org/view/15918-Transformation.html
- http://www.dnalc.org/view/15916-DNA-transformation.html
Exercise: Transformation of Bacteria with RE Identified Plasmids
- Retrieve miniprepped plasmid from freezer and allow to thaw on ice.
- Bring 2 agar plates to room temperature
- 1 plate will contain antibiotic, X-Gal and arabinose
- 1 plate will contain only antibiotic
- Pipette 250μl of transformation buffer (20mM CaCl2) into a microfuge tube onto ice for 10 minutes
- Take an inoculating loop and remove a single colony of bacteria from a freshly streaked plate grown overnight
- Swirl bacteria in tube with transforming solution to distribute bacteria throughout solution
- Pipette 5 μl of plasmid into the tube and incubate on ice for 10 minutes
- During this incubation, flip the warmed plates and label them with your group names. Note which one has X-Gal and Arabinose.
- Place transformation tubes into 42°C heatblock for 1 minute to heat shock the cells
- Add 250μl fresh LB and incubate at 37°C for 15 minutes.
- Pipette 200μl of transformation solution onto each plate and spread across the plate.
- Turn plates agar side up and place them into 37°C incubator overnight. (your instructor will retrieve them and place them into refrigerator)
Hypothesize: What will I expect of my transformed cells?
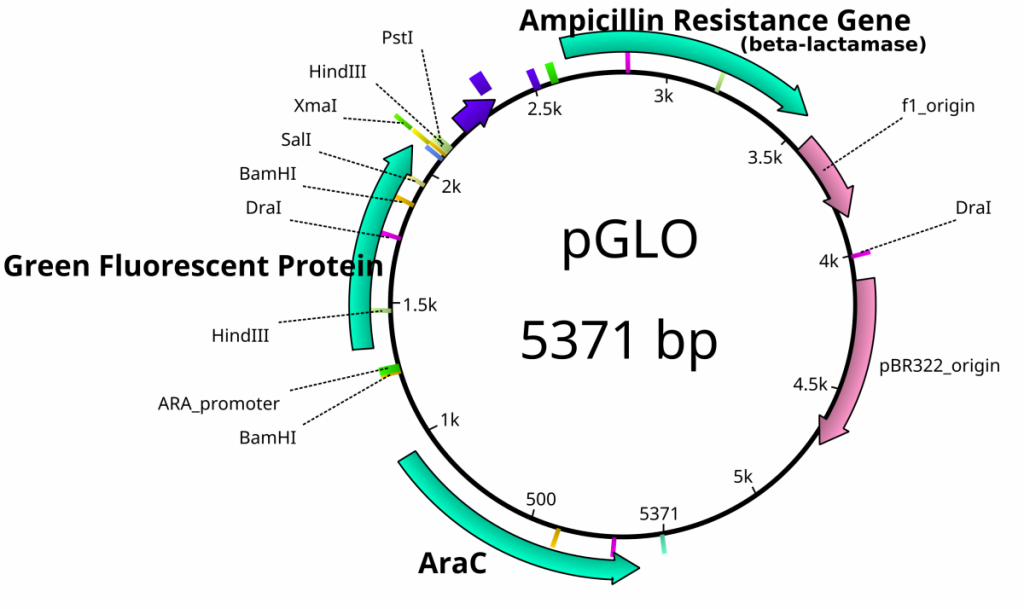
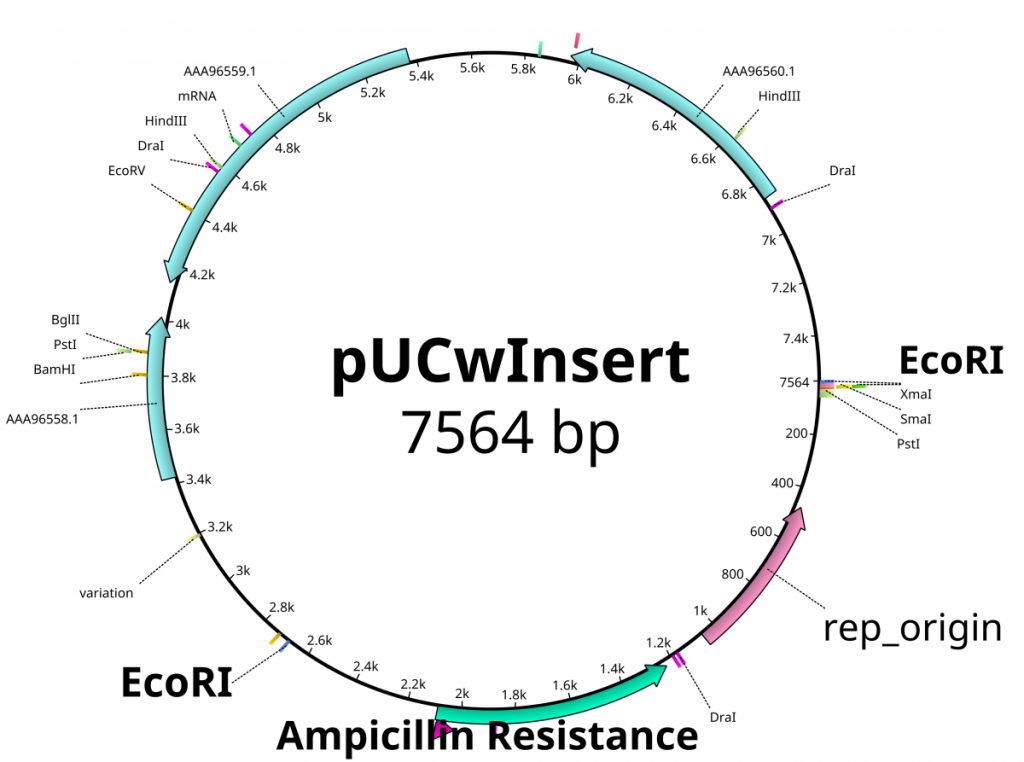
From the previous lab, we can identify our plasmids. The plasmids are either pGlo, pUC18/19 or pUC18/19 with a 6kb insert disrupting the LacZ gene. pGlo contains a gene that encodes the protein GFP that will fluoresce green under UV light and is 5.4kb. pUC is typically 2.7kb in size. LacZ is a gene encoding the protein β-Galactosidase, the enzyme that hydrolyzes lactose into the monosaccharides galactose and glucose. X-Gal is a chemical resembling lactose, however upon hydrolysis, the molecule deposits a blue coloring into the cell.
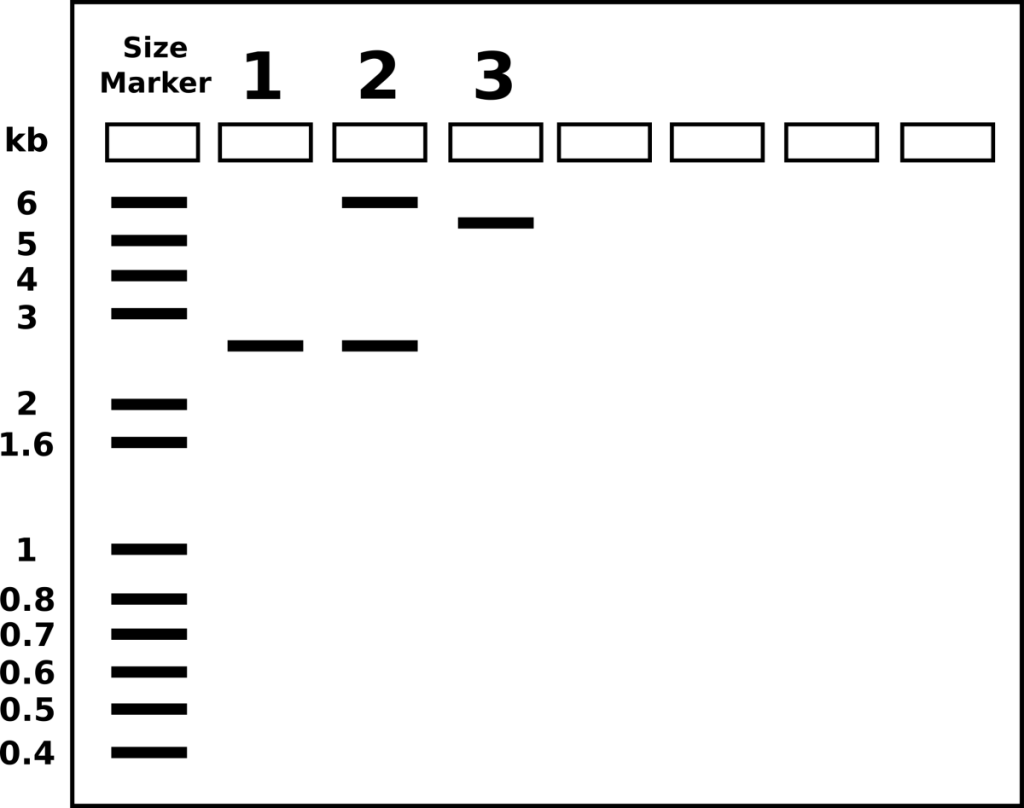
- If the previously mentioned plasmids were digested by EcoRI, label the lanes below with the appropriate plasmid (pGlo, pUC, pUC-inserted)
- Predict if your transformants will be green under UV, white in all conditions or blue.
- For additional help on this problem, utilize the In silico digestion activity